Help Information
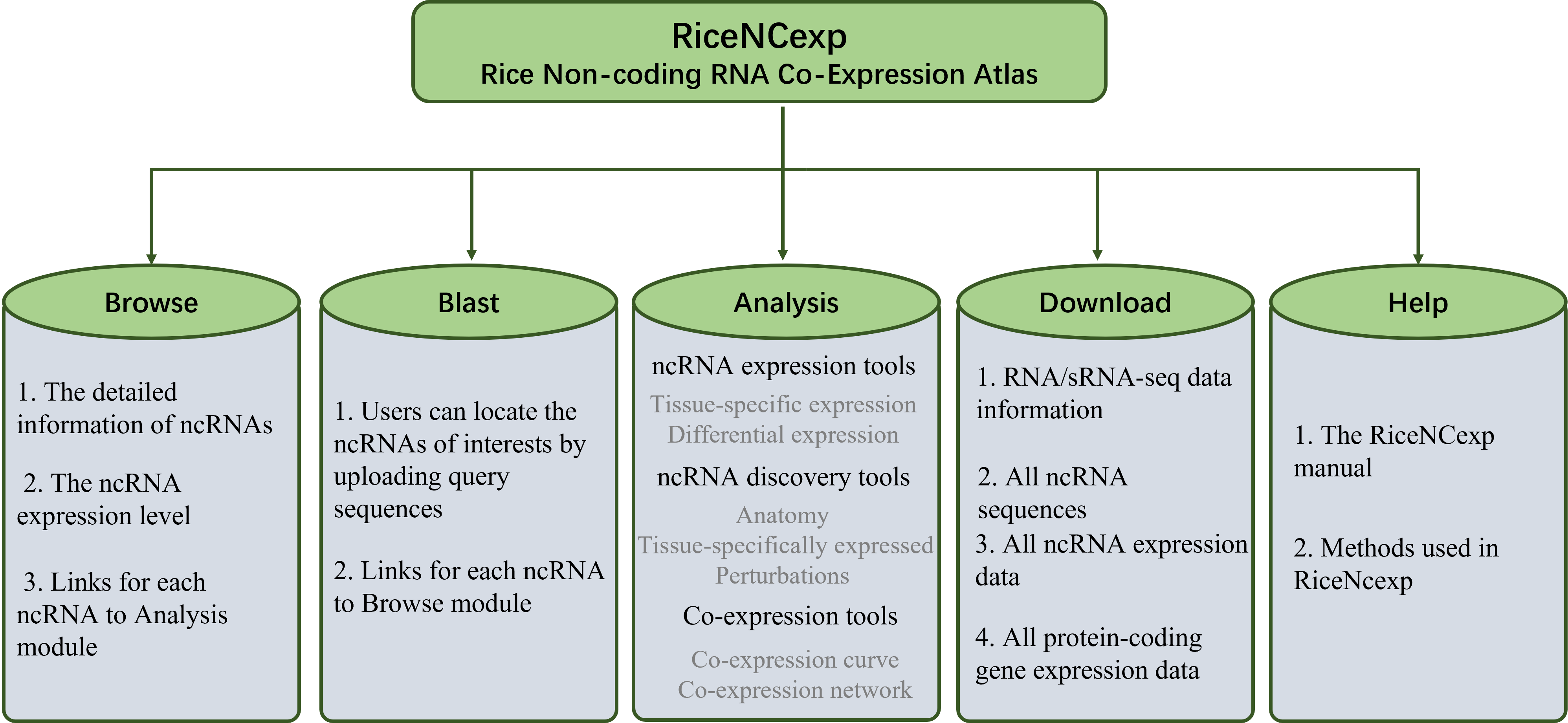
This module shows some basic information of the ncRNAs(microRNAs, phasiRNAs, lncRNAs and PHAS gene) used in this atlas.
"DE" is short for differential expression. (log2(fold change) > 1 or log2(fold change) < 1 and FDR < 0.01)
"TSE" is short for tissue-specific expression. (tau > 0.9 and Average expression(tpm) in the tissue > 1)
"Y" denotes featured gene/sRNA under a certain context
"N" denotes not a featured gene/sRNA under the all context
MicroRNAs model:
We combined the mature miRNA sequences of miRBase and PmiREN and used them as the basic information of miRNA in our atlas. If the miRNA sequence only exists in the miRBase database, the suffix of the microRNA name is "_miRbase". If the sequence only exists in PmiREN database, the suffix of the microRNA name is "_PmiREN". If the sequence exists in both miRBase and PmiREN, the suffix of the microRNA name is "_Both".In addition, Besides,we prediected the novel miRNAs across 307 sRNA-seq datasets by miRDeep-P2 and suffixed them with "_RiceNCexp".
col 1: The name of the miRNA.
col 2: The source of the miRNA (miRbase / PmiREN / Both / RiceNCexp).
col 3: The confidence of the miRNA.
col 4: The location of the miRNA.
col 5: The start position of the miRNA on chromosome.
col 6: The end position of the miRNA on chromosome.
col 7: The strand of the miRNA on chromosome.
col 8: The expression feature of the miRNA.
col 9: Show the detail of the miRNA.
Phased siRNAs(phasiRNAs) model:
col 1: The name of the phasiRNA.
col 2: The source(PHAS gene) of the phasiRNA.
col 3: The strand of the phasiRNA on PHAS gene.
col 4: The strart position of the phasiRNA on PHAS gene.
col 5: The NCBI symbol of the PHAS gene.
col 6: The location of the phasiRNA.
col 7: The strand of the phasiRNA on chromosome.
col 8: The annotation of the PHAS gene.
col 9: The expression feature of the phasiRNA.
col 10: Show the detail of the phasiRNA.
lncRNAs model:
We obtained 7898 novel rice lncRNAs based on transcript assembly. The names of these novel lncRNAs are prefixed with "OSA-LNCN".
Based on the genomic locations in respect to protein-coding genes, we classified lncRNAs into seven groups: Intergenic, Intronic (S), Intronic (AS), Overlapping (S), Overlapping (AS), Sense, and Antisense. 'S' in the bracket represents that lncRNAs are in the same strand of protein-coding RNAs, and 'AS' represents that lncRNAs are in the antisense strand of protein-coding RNAs.
Seven classification:
Intergenic: lncRNAs are transcribed from intergenic regions;
Intronic (S): lncRNAs are transcribed entirely from introns of protein-coding genes;
Intronic (AS): lncRNAs are transcribed from antisense strand of protein-coding genes and the entire sequences are covered by introns of protein-coding genes;
Overlapping (S): lncRNAs that contain coding genes within an intron on the sense strand;
Overlapping (AS): lncRNAs that contain coding genes within an intron on the antisense strand;
Sense: lncRNAs are transcribed from the sense strand of protein-coding genes and the entire sequence of lncRNAs are covered by protein-coding genes (Intronic lncRNAs are not included), or the entire sequence of protein-coding genes are covered by lncRNAs (Overlapping lncRNAs are not included), or both lncRNAs and protein-coding genes intersect each other partially;
Antisense: lncRNAs are transcribed from the antisense strand of protein-coding genes and the entire sequence of lncRNAs are covered by protein-coding genes (Intronic lncRNAs are not included), or the entire sequence of protein-coding genes are covered by lncRNAs (Overlapping lncRNAs are not included), or both lncRNAs and protein-coding genes intersect each other partially.
col 1: The name of the lncRNA.
col 2: The symbol of the lncRNA.
col 3: The classification of the lncRNA.
col 4: The location of the lncRNA.
col 5: The start position of the lncRNA on chromosome.
col 6: The end position of the lncRNA on chromosome.
col 7: The strand of the lncRNA on chromosome.
col 8: The expression feature of the lncRNA.
col 9: Show the detail of the lncRNA.
PHAS Gene model:
col 1: The name of the PHAS gene.
col 2: The symbol of the PHAS gene.
col 3: The location of the PHAS gene.
col 4: The start position of the PHAS gene on chromosome.
col 5: The end position of the PHAS gene on chromosome.
col 6: The strand of the PHAS gene on chromosome.
col 7: The expression feature of the PHAS gene.
col 8: Show the detail of the PHAS gene.
Example:
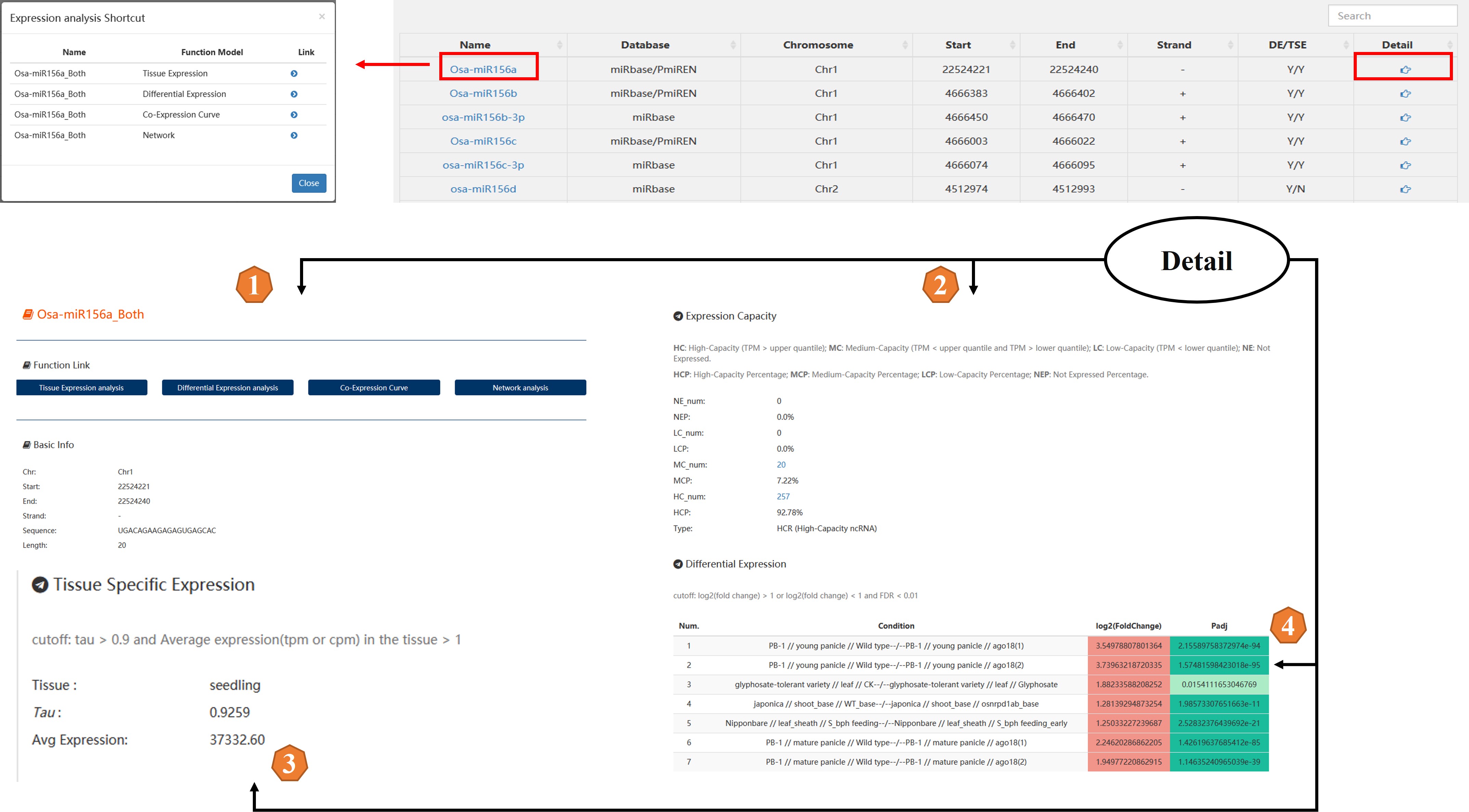
Detailed description of the above section:
A. Function entrance of RiceNCexp.
B. To ensure the web page loading speed, each page displays 20 records by default. If you want to fuzzy search all miRNAs information by the search box, you need to select "all". But this will sacrifice a little bit of page loading speed.
C. Detail:
1. The function entrance of the ncRNA including tissue-specific expression analysis, differential expression analysis, co-expression curve and network analysis.
2. The expression level of the ncRNA.
3. If the ncRNA is a tissue-specific expression ncRNA, the tissue-specific expression information will be displayed here.
4. If the ncRNA is a differential expression ncRNA, the differential expression information will be displayed here.
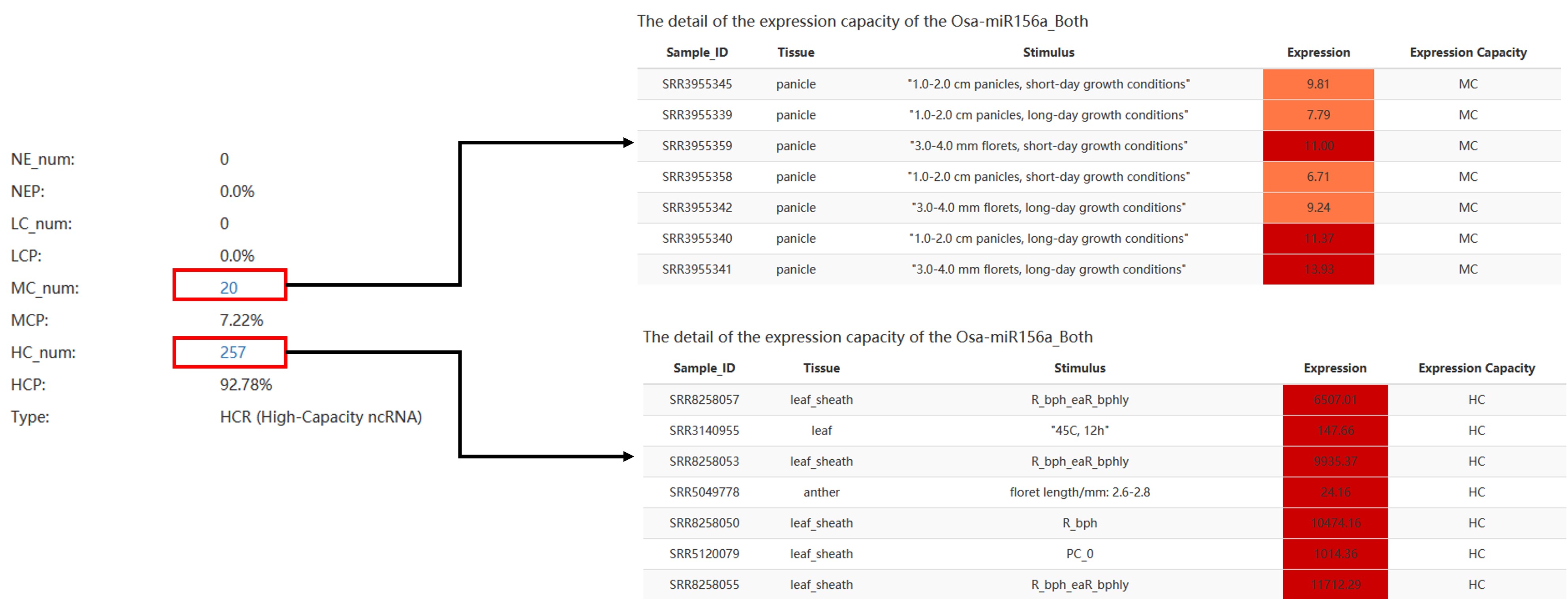
Expression Capacity Model:
The High-Expressed (HE) means that the expression of the ncRNA is greater than the upper quartile in a certain sample.
The Medium-Expressed (ME) means that the expression of the ncRNA is between the upper quartile and the lower quartile in a certain sample.
The Low-Expressed (LE) means that the expression of the ncRNA is less than the lower quartile in a certain sample.
HEP: High-Expressed Percentage; MEP: Medium-Expressed Percentage; LEP: Low-Expressed Percentage; NEP: Not Expressed Percentage.
col 1: The name of ncRNA.
col 2: The number of NE samples.
col 3: The percentage of NE samples.
col 4: The number of LE samples.
col 5: The percentage of LE samples.
col 6: The number of ME samples.
col 7: The percentage of ME samples.
col 8: The number of HE samples.
col 9: The percentage of HE samples.
col 10: The type(HE/ME/LE) of ncRNA.
Example:
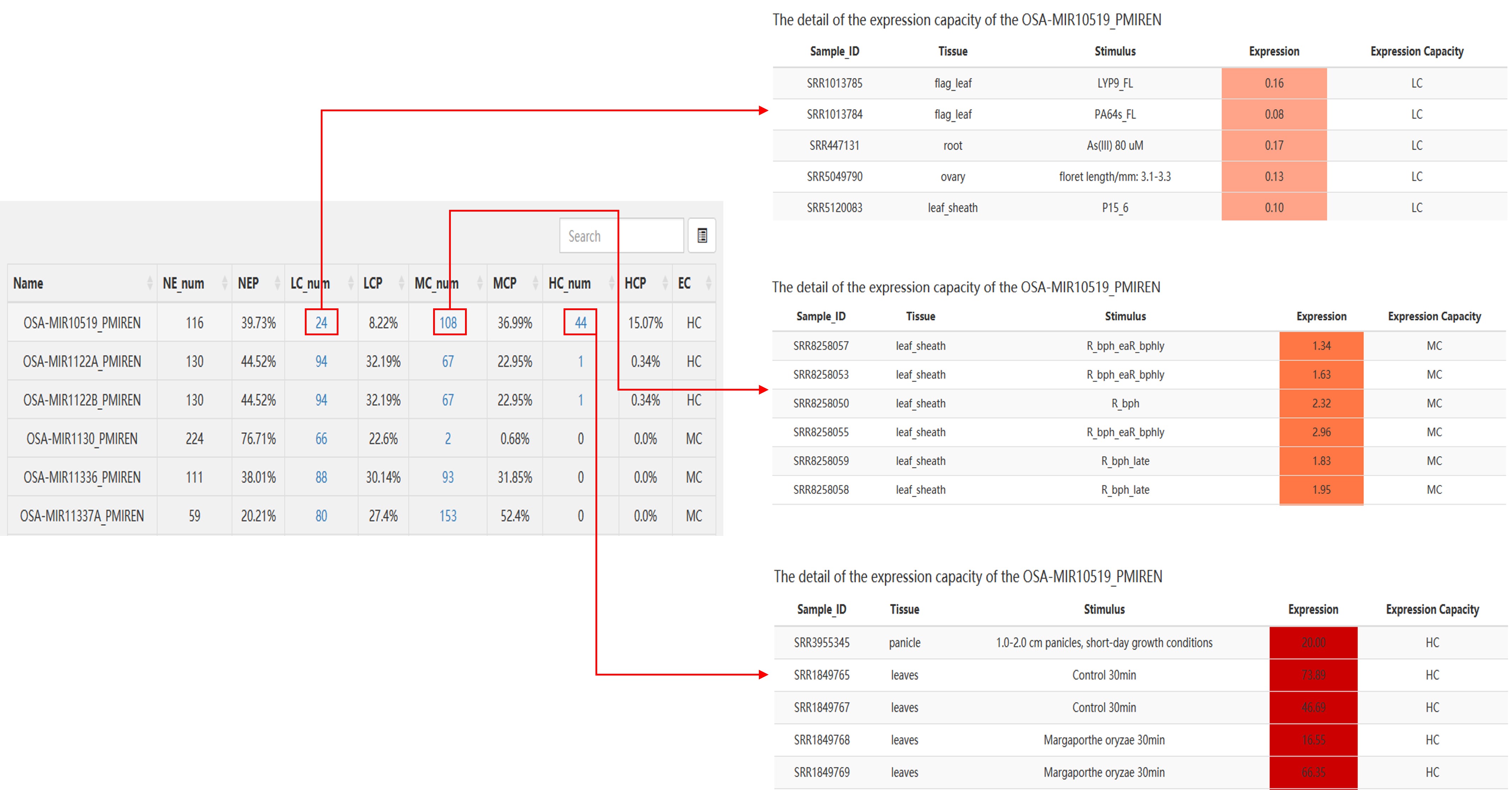
Detailed description of the above section:
The color gradient represents the amount of expression
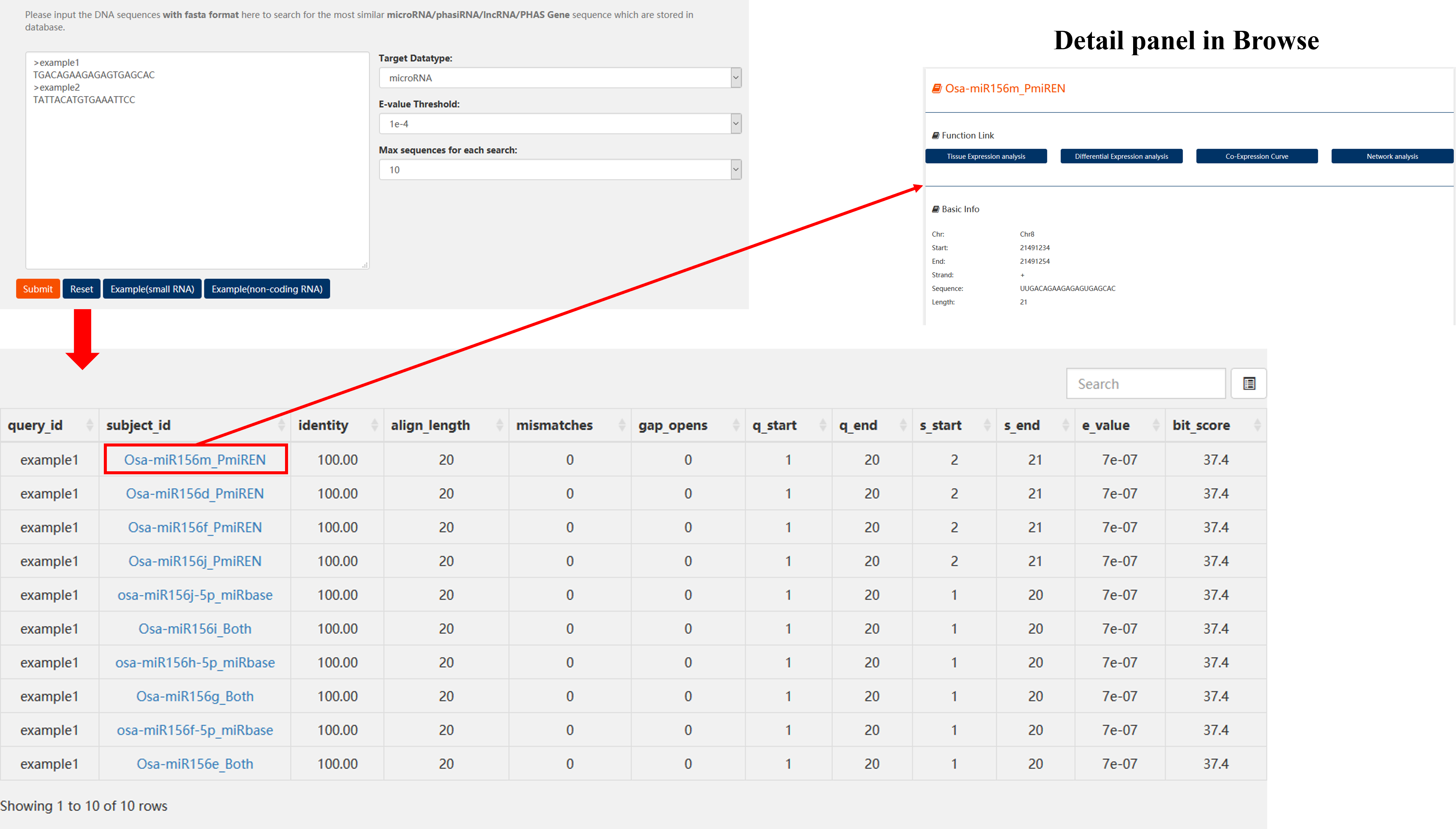
Input a DNA sequence with fasta format to search for the most similar microRNA/phasiRNA/lncRNA/PHAS Gene sequence which are stored in our altas.
microRNA/phasiRNA last Results:
col 1: The id of the input sequence
col 2: The id of the matched sRNA
col 3: Alignment percentage
col 4: Alignment length
col 5: Number of mismatches
col 6: Number of gap openings
col 7: Start of alignment in query
col 8: End of alignment in query
col 9: Start of alignment in subject
col 10: End of alignment in subject
col 11: Expect value
col 12: Bit score
lncRNA/PHAS Gene Blast Results:
col 1: The id of the input sequence
col 2: The id of the matched ncRNA
col 3: the symbol of the matched ncRNA
col 4: the name of the matched ncRNA in the altas
col 5: Alignment percentage
col 6: Alignment length
col 7: Number of mismatches
col 8: Number of gap openings
col 9: Start of alignment in query
col 10: End of alignment in query
col 11: Start of alignment in subject
col 12: End of alignment in subject
col 13: Expect value
col 14: Bit score
Example:
There are two basic functions in this function block.(Tissue-specific Expression, Differential Expression)
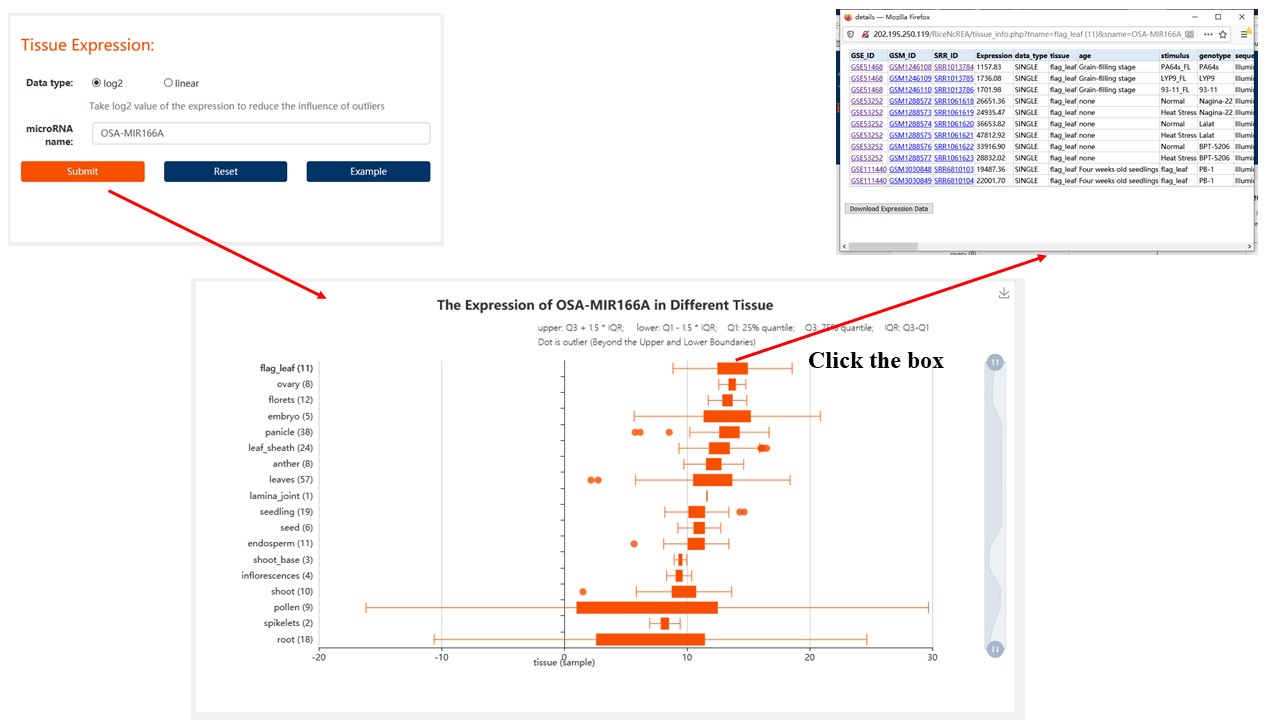
1. Tissue-specific Expression
Find the expression of non-coding RNA you are interested in under rice tissues.
For miRNA/phasiRNA data, we have 19 different rice tissues.
For lncRNA/PHAS Gene data, we have 22 different rice tissues.
Data type:
linear: For miRNA/phasiRNA data, we use CPM to represent their expression; For lncRNA/PHAS Gene data, we use TPM to represent their expression
log2: Take log2 value for ncRNA expression. (log2(Exp+1)).
Result:
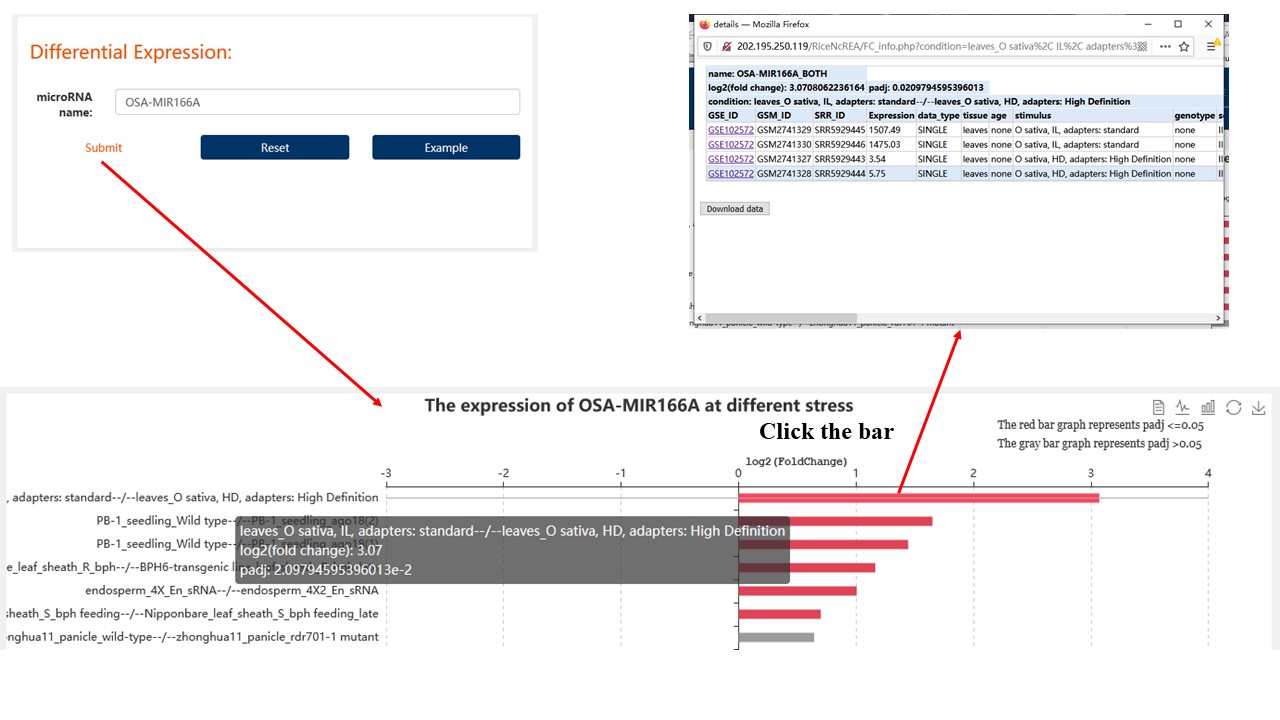
2. Differential Expression
Find the differential expression of the non-coding RNA you are interested in under each stress.
For miRNA/phasiRNA data, we have 32 different stress conditions.
For lncRNA/PHAS Gene data, we have 78 different stress conditions.
Label format:
genotype//tissue//condition (control)--/--(treatment) genotype//tissue//condition
Result:
There are three basic functions in this function block.(Tissue specifically expressed, Anatomy, Perturbations)
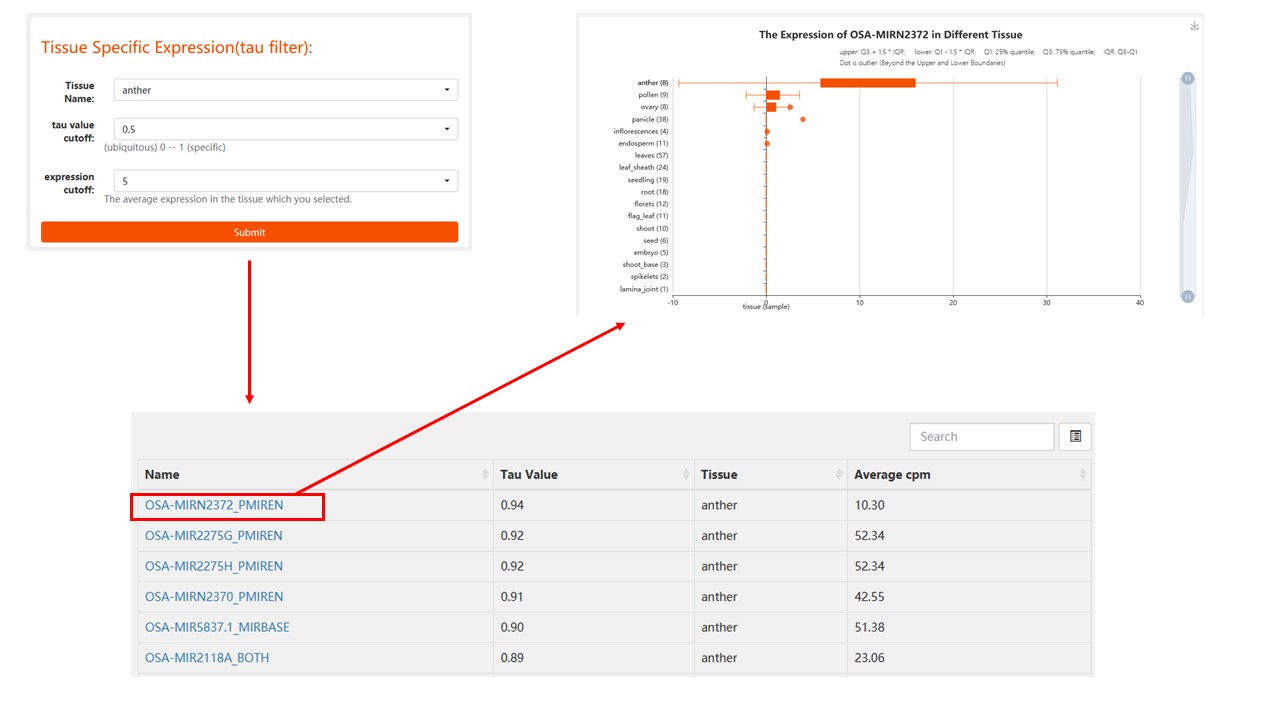
1. Tissue specifically expressed
Find the specific expression of non-coding RNA in the rice tissues you are interested in. (determined by tau value)
For tau value, the ncRNA tends to be widely expressed in various tissue if it is close to 0, while when tau is closer to 1, ncRNA is more predominantly expressed in certain tissue/organ.
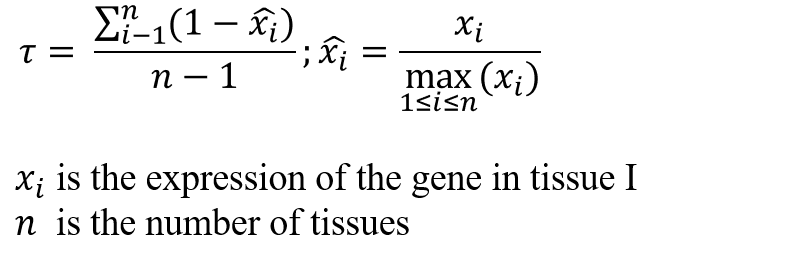
Result:
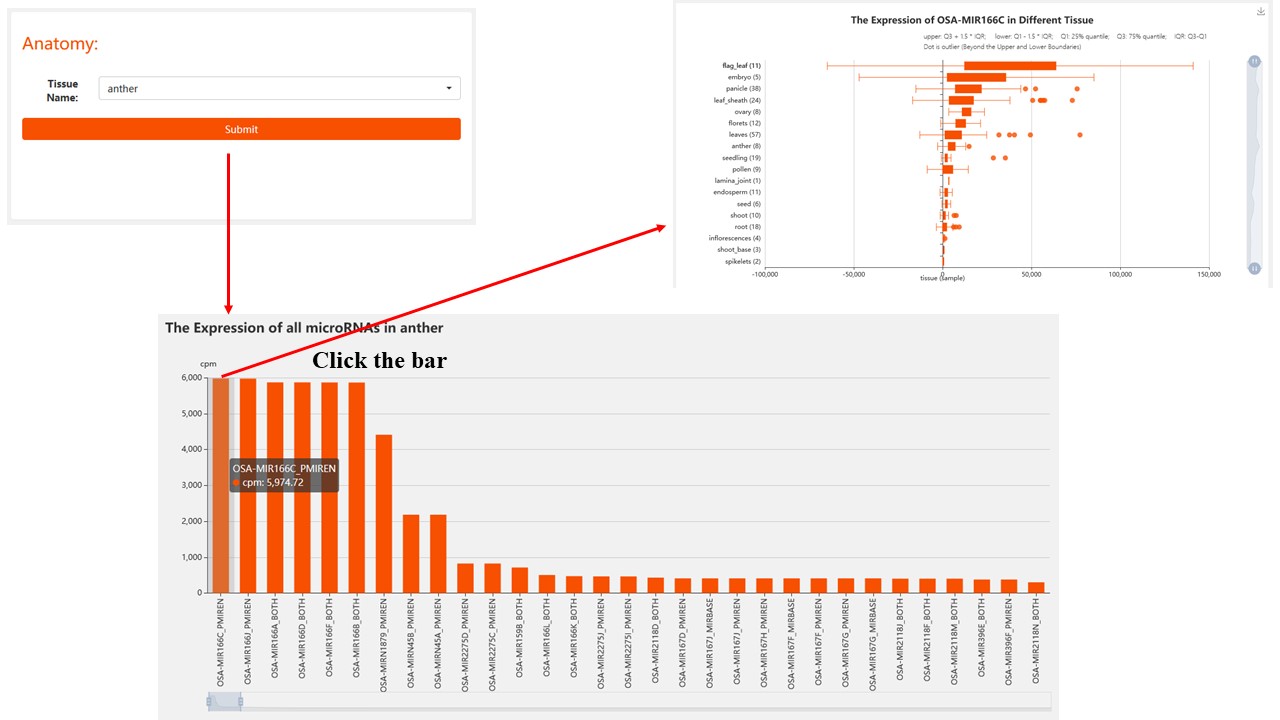
2. Anatomy
Find the expression distribution of non-coding RNA in the rice tissues you are interested in.
For miRNA/phasiRNA data, we have 19 different rice tissues.
For lncRNA/PHAS Gene data, we have 22 different rice tissues.
Result:
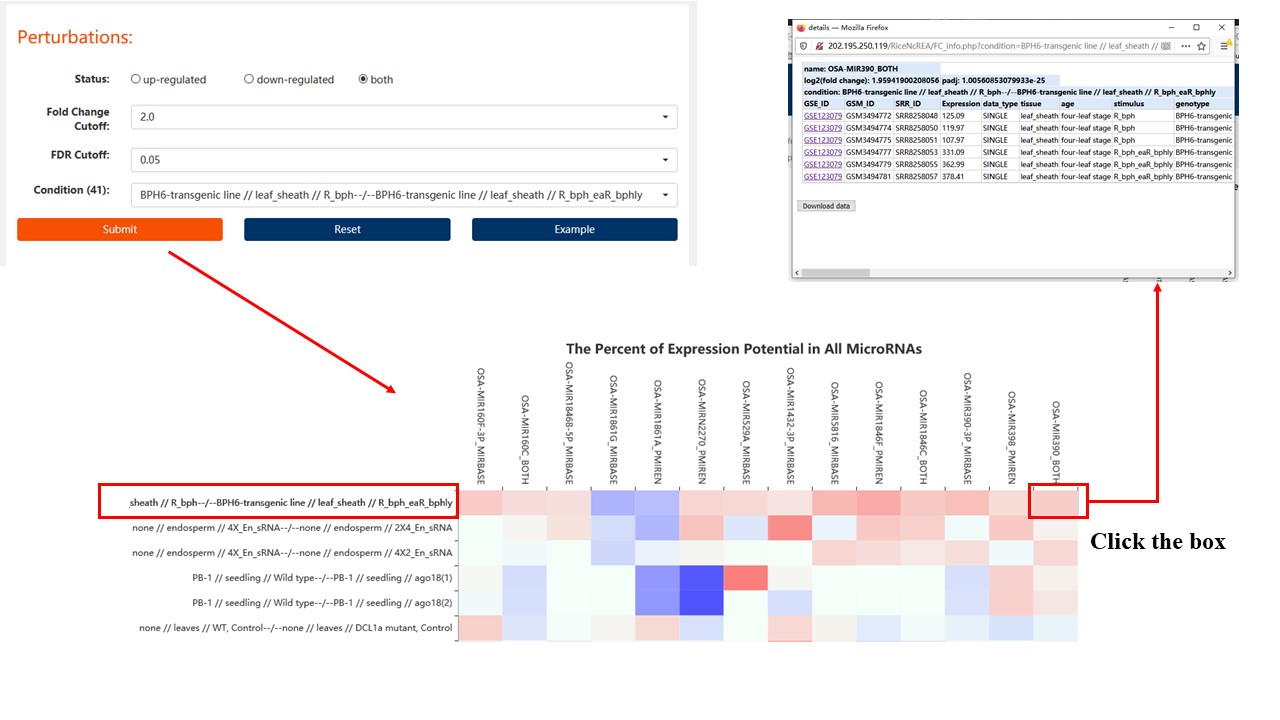
3. Perturbations
Find the differential expression of non-coding RNA in the stress conditions you are interested in.
Label format:
genotype//tissue//condition (control)--/--(treatment) genotype//tissue//condition
Result:
The x-axis of the result represents the name of ncRNA that meets the parameter(status, fold change cutoff, FDR cutoff and the condition) which you choosed. The first line of the heat map is the condition which you choosed. The following is the differential expression of these ncRNAs in other conditions.
Co-expression
There are two basic functions in this function block.(Co-expression curve, Network)
1. Co-Expression Curve
This is a function module for online calculation of co-expression relationship based on Pearson correlation coefficient (PCC). The co-Expression curve module contains two query functions: One to Many and One to One
One to Many: Query all co-expressed genes/ncRNAs of a certain ncRNA/gene under given parameters.
One to One: Query the co-expression correlation coefficient (PCC) between specific two ncRNA/gene.
Parameter:
Source Data Type: Five target types can be selected, including four non-coding RNAs in RiceNCexp, and the protein-coding RNAs (msu7).
Target Data Type: Five target types can be selected, including four non-coding RNAs in RiceNCexp, and the portein-coding RNAs (msu7).
Library Type: We used both RNA-seq sequencing data and sRNA-seq sequencing data from the same experiments as co-expression libraries, and divided libraries into two categories, one is different tissues, a total of 11 libraries, the other is all datasets, including all kinds of rice tissues and stresses, a total of 116 libraries.
Correlation: Positive: Positive correlation; Negative: Negative correlation
PCC cutoff: Threshold of Pearson correlation coefficient(PCC)
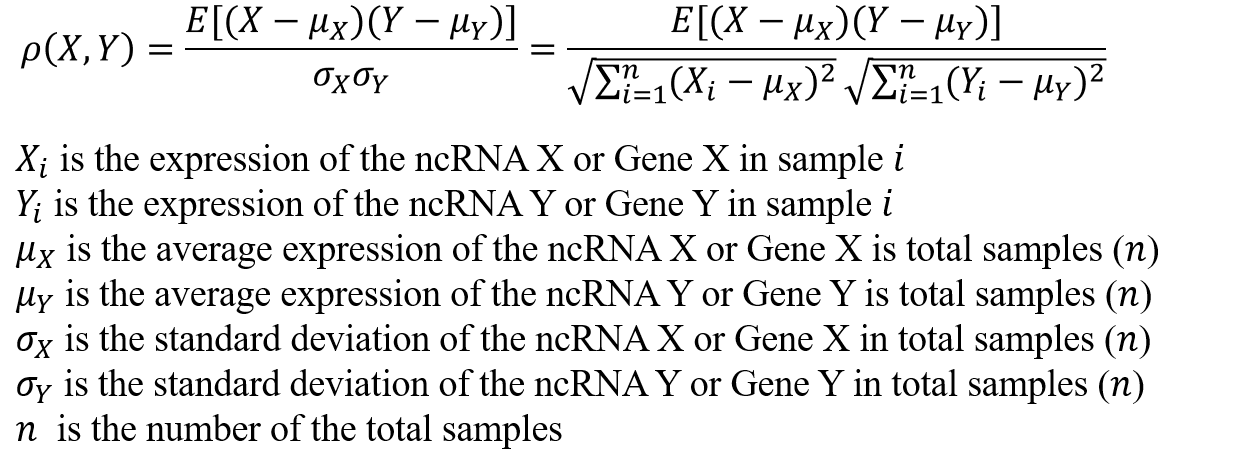
Result:
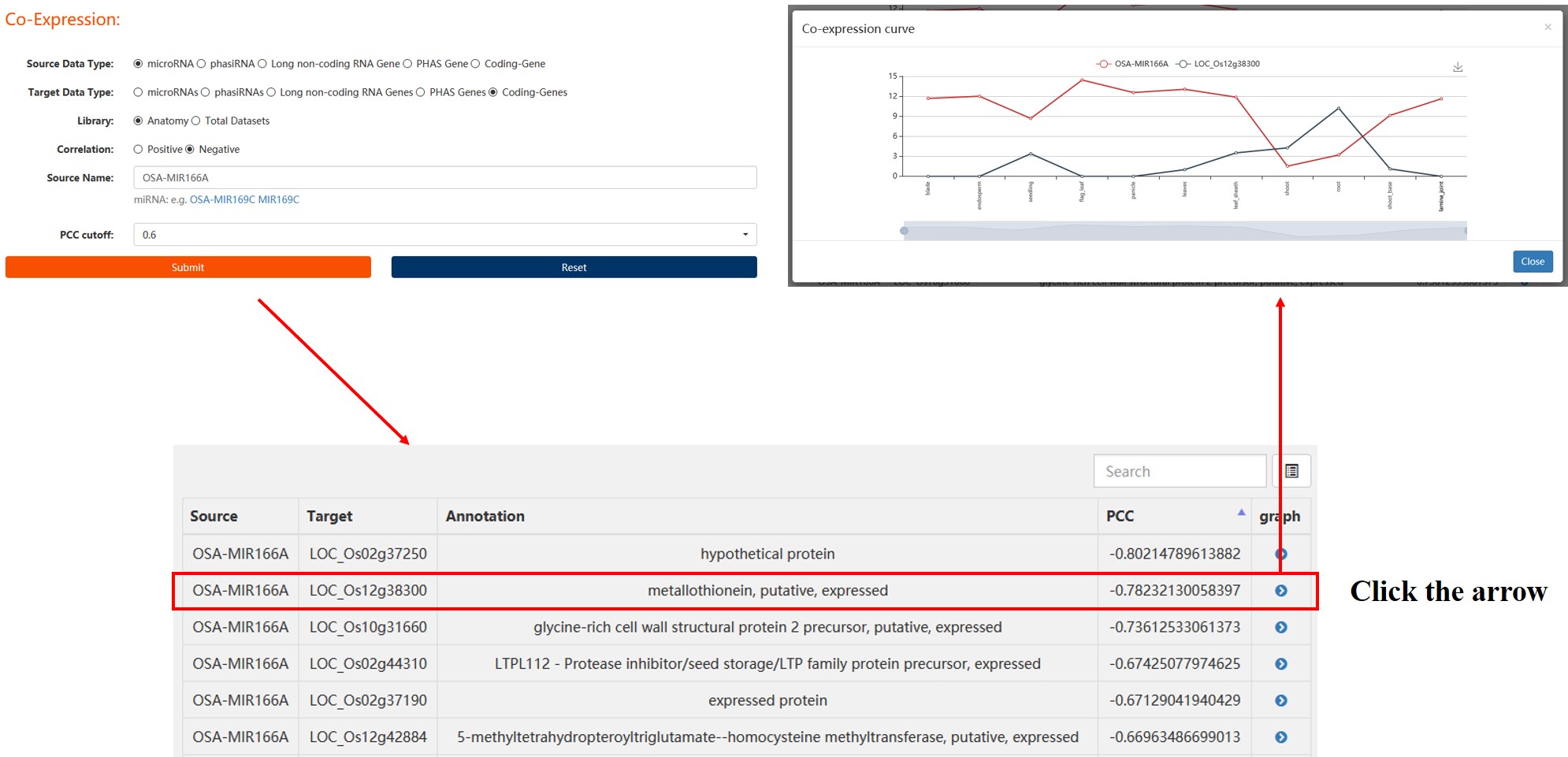
2. Co-Expression Network
This is a function module for online analysis of the complexity of co-expression-dependent network.
Parameter:
Start Point Type: Five target types can be selected, including four non-coding RNAs in RiceNCexp, and the protein-coding RNAs (msu7).
Library Type: We used both RNA-seq sequencing data and sRNA-seq sequencing data from the same experiments as co-expression libraries, and divided libraries into two categories, one is different tissues, a total of 11 libraries, the other is all datasets, including all kinds of rice tissues and stresses, a total of 116 libraries.
Layers: Number of network layers (Maximum 3 layers).
e.g. Two Layers network: if A is co-expressed with B, B is co-expressed with C, A may be co-expressed with C.
The larger the number of layers, the slower the analysis speed.
PCC cutoff: Threshold of Pearson correlation coefficient(PCC) in each layer.
The larger the PCC, the faster the analysis speed.
maximum of nodes: Maximum number of nodes in each layer.
e.g. the maximum of nodes is 10. In this layer of network, the top 10 co-expression pairs with strong correlation are displayed in the network.
The larger the number of nodes, the slower the analysis speed.
Start Point(s): The root node. (The name of the input non-coding RNAs or coding genes)
One query per line.
The query cannot exceed 10 lines, otherwise the running speed is too slow and the generated co-expression network is too complicated.
Result:
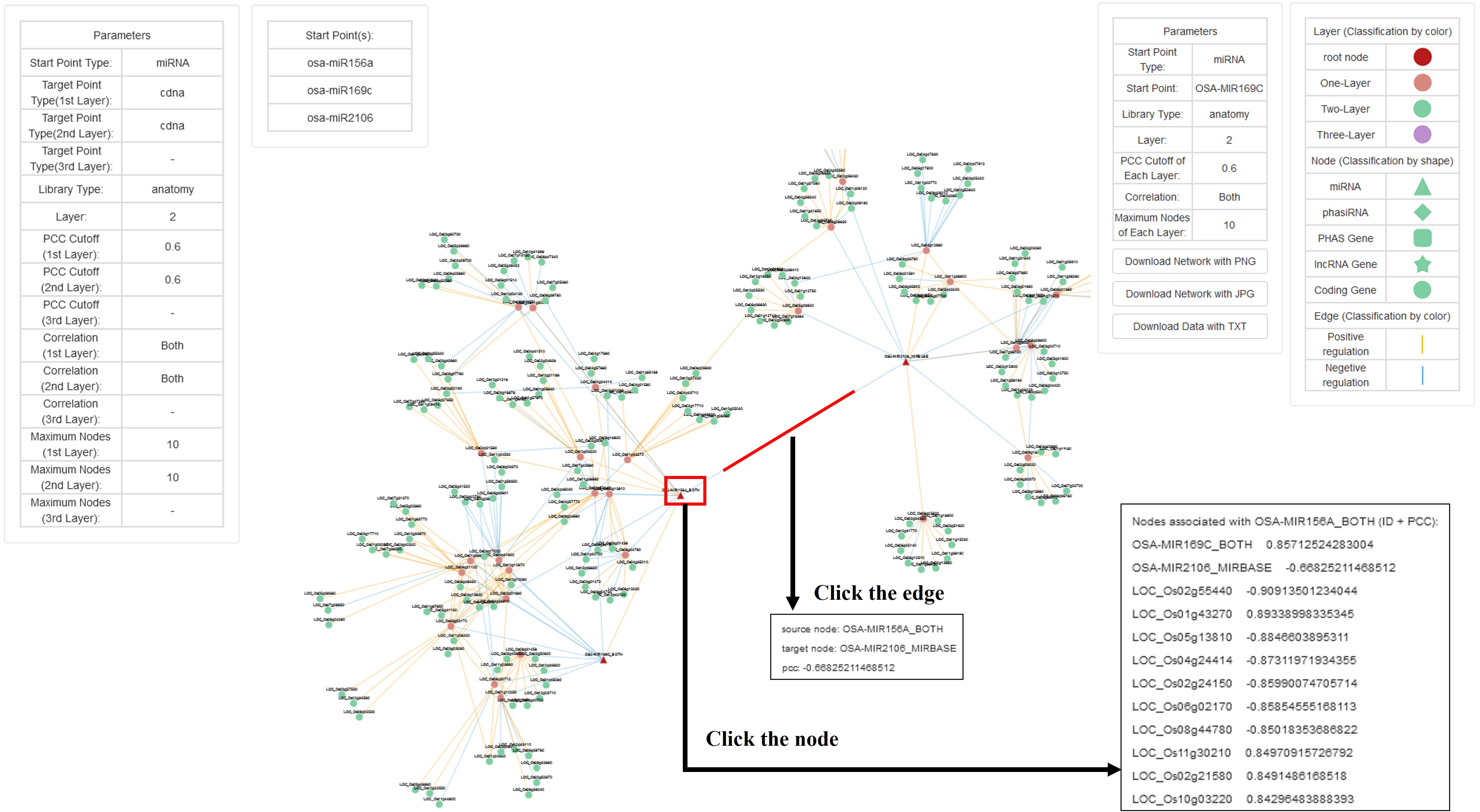
Filter conditions for low-expressed genes:
For 11 libraries (anatomy): The gene/ncRNA is expressed in at least 3 libraries.
For 116 libraries (total datasets): The gene/ncRNA is expressed in at least 10 libraries.
| Tissue | Samples |
|---|---|
| lamina_joint | SRR12416717,SRR12416718,SRR12416719,SRR12416720,SRR12416721,SRR12416722,SRR12416723,SRR12416724,SRR12416725,SRR12416726,SRR976168,SRR976169,SRR976170 |
| blade | SRR12416727,SRR12416728,SRR12416729,SRR12416710,SRR12416711,SRR12416712,SRR12416713,SRR12416714,SRR12416715,SRR12416716 |
| leaf | SRR10640869,SRR10640870,SRR7058086,SRR7058087,SRR7058088,SRR7058089,SRR6498704,SRR6498705,SRR6498706,SRR6498707,SRR6498708,SRR6498709,SRR6498710,SRR6498711,SRR4098199,SRR3203681,SRR3203682,SRR3203683,SRR2862232,SRR2862233,SRR2862234,SRR2862235,SRR2862236,SRR2862237,SRR3130386,SRR3130387,SRR3130388,SRR3130389,SRR3130390,SRR3130391,SRR3130392,SRR3130393,SRR3130394,SRR3130395 |
| shoot_base | SRR9070097,SRR9070098,SRR9070099,SRR9070106,SRR9070107,SRR9070108 |
| leaf_sheath | SRR8247767,SRR8247768,SRR8247769,SRR8247770,SRR8247771,SRR8247772,SRR8247773,SRR8247774,SRR8247775,SRR8247776,SRR8247777,SRR8247778,SRR8247779,SRR8247780,SRR8247781,SRR8247782,SRR8247783,SRR8247784 |
| panicle | SRR5940080,SRR5940083,SRR4098193,SRR6807850,SRR6807851,SRR1013793,SRR1013794,SRR1013795 |
| endosperm | SRR1743007,SRR1743008,SRR1743009,SRR1743010,SRR1743011,SRR1743012,SRR1743013,SRR1743014,SRR1743015 |
| seedling | SRR4098173,SRR4098176,SRR4098179,SRR4098187,SRR4098190 |
| root | SRR4098196,SRR447117,SRR447120 |
| flag_leaf | SRR6807844,SRR6807845,SRR1013790,SRR1013791,SRR1013792 |
| shoot | SRR447122,SRR447123,SRR447124,SRR447125,SRR447126 |
| Num | Sample ID | Data Type | Tissue | Age | Stimulus | Genotype |
|---|---|---|---|---|---|---|
| 1 | SRR12416717 | PAIRED | lamina_joint | developmental stage: S1 | mRNA_LJ_S1 | none |
| 2 | SRR12416718 | PAIRED | lamina_joint | developmental stage: S1 | mRNA_LJ_S1 | none |
| 3 | SRR12416719 | PAIRED | lamina_joint | developmental stage: S2 | mRNA_LJ_S2 | none |
| 4 | SRR12416720 | PAIRED | lamina_joint | developmental stage: S2 | mRNA_LJ_S2 | none |
| 5 | SRR12416721 | PAIRED | lamina_joint | developmental stage: S3 | mRNA_LJ_S3 | none |
| 6 | SRR12416722 | PAIRED | lamina_joint | developmental stage: S3 | mRNA_LJ_S3 | none |
| 7 | SRR12416723 | PAIRED | lamina_joint | developmental stage: S4 | mRNA_LJ_S4 | none |
| 8 | SRR12416724 | PAIRED | lamina_joint | developmental stage: S4 | mRNA_LJ_S4 | none |
| 9 | SRR12416725 | PAIRED | lamina_joint | developmental stage: S5 | mRNA_LJ_S5 | none |
| 10 | SRR12416726 | PAIRED | lamina_joint | developmental stage: S5 | mRNA_LJ_S5 | none |
| 11 | SRR12416727 | PAIRED | blade | developmental stage: S1 | mRNA_Blade_S1 | none |
| 12 | SRR12416728 | PAIRED | blade | developmental stage: S1 | mRNA_Blade_S1 | none |
| 13 | SRR12416729 | PAIRED | blade | developmental stage: S2 | mRNA_Blade_S2 | none |
| 14 | SRR12416710 | PAIRED | blade | developmental stage: S2 | mRNA_Blade_S2 | none |
| 15 | SRR12416711 | PAIRED | blade | developmental stage: S3 | mRNA_Blade_S3 | none |
| 16 | SRR12416712 | PAIRED | blade | developmental stage: S3 | mRNA_Blade_S3 | none |
| 17 | SRR12416713 | PAIRED | blade | developmental stage: S4 | mRNA_Blade_S4 | none |
| 18 | SRR12416714 | PAIRED | blade | developmental stage: S4 | mRNA_Blade_S4 | none |
| 19 | SRR12416715 | PAIRED | blade | developmental stage: S5 | mRNA_Blade_S5 | none |
| 20 | SRR12416716 | PAIRED | blade | developmental stage: S5 | mRNA_Blade_S5 | none |
| 21 | SRR10640869 | SINGLE | leaf | at the tillering stage | IR24-0h | IR24 |
| 22 | SRR10640870 | SINGLE | leaf | at the tillering stage | IRBB5-0h | IRBB5 |
| 23 | SRR9070097 | PAIRED | shoot_base | 4-week | WT_base_mRNA180209 | japonica |
| 24 | SRR9070098 | PAIRED | shoot_base | 4-week | WT_base_mRNA180209 | japonica |
| 25 | SRR9070099 | PAIRED | shoot_base | 4-week | WT_base_mRNA180209 | japonica |
| 26 | SRR9070106 | PAIRED | shoot_base | 4-week | osnrpd1ab_base_mRNA180209 | japonica |
| 27 | SRR9070107 | PAIRED | shoot_base | 4-week | osnrpd1ab_base_mRNA180209 | japonica |
| 28 | SRR9070108 | PAIRED | shoot_base | 4-week | osnrpd1ab_base_mRNA180209 | japonica |
| 29 | SRR8247767 | PAIRED | leaf_sheath | four-leaf stage | R-0 | Nipponbare |
| 30 | SRR8247768 | PAIRED | leaf_sheath | four-leaf stage | R-0 | Nipponbare |
| 31 | SRR8247769 | PAIRED | leaf_sheath | four-leaf stage | R-0 | Nipponbare |
| 32 | SRR8247770 | PAIRED | leaf_sheath | four-leaf stage | R-early | Nipponbare |
| 33 | SRR8247771 | PAIRED | leaf_sheath | four-leaf stage | R-early | Nipponbare |
| 34 | SRR8247772 | PAIRED | leaf_sheath | four-leaf stage | R-early | Nipponbare |
| 35 | SRR8247773 | PAIRED | leaf_sheath | four-leaf stage | R-late | Nipponbare |
| 36 | SRR8247774 | PAIRED | leaf_sheath | four-leaf stage | R-late | Nipponbare |
| 37 | SRR8247775 | PAIRED | leaf_sheath | four-leaf stage | R-late | Nipponbare |
| 38 | SRR8247776 | PAIRED | leaf_sheath | four-leaf stage | S-0 | Nipponbare |
| 39 | SRR8247777 | PAIRED | leaf_sheath | four-leaf stage | S-0 | Nipponbare |
| 40 | SRR8247778 | PAIRED | leaf_sheath | four-leaf stage | S-0 | Nipponbare |
| 41 | SRR8247779 | PAIRED | leaf_sheath | four-leaf stage | S-eaSly | Nipponbare |
| 42 | SRR8247780 | PAIRED | leaf_sheath | four-leaf stage | S-eaSly | Nipponbare |
| 43 | SRR8247781 | PAIRED | leaf_sheath | four-leaf stage | S-eaSly | Nipponbare |
| 44 | SRR8247782 | PAIRED | leaf_sheath | four-leaf stage | S-late | Nipponbare |
| 45 | SRR8247783 | PAIRED | leaf_sheath | four-leaf stage | S-late | Nipponbare |
| 46 | SRR8247784 | PAIRED | leaf_sheath | four-leaf stage | S-late | Nipponbare |
| 47 | SRR5940080 | PAIRED | panicle | young panicles before heading date | Wild type | none |
| 48 | SRR5940083 | PAIRED | panicle | young panicles before heading date | dcl3b knockdown mutant | none |
| 49 | SRR7058086 | PAIRED | leaf | treatment time: 20d | rice_mock | Nipponbare |
| 50 | SRR7058087 | PAIRED | leaf | treatment time: 20d | rice_mock | Nipponbare |
| 51 | SRR7058088 | PAIRED | leaf | treatment time: 20d | rice_RSV | Nipponbare |
| 52 | SRR7058089 | PAIRED | leaf | treatment time: 20d | rice_RSV | Nipponbare |
| 53 | SRR6498704 | SINGLE | leaf | 3 weeks old | "DCL1a mutant, Control" | none |
| 54 | SRR6498705 | SINGLE | leaf | 3 weeks old | "DCL1a mutant, M. oryzae Infected" | none |
| 55 | SRR6498706 | SINGLE | leaf | 3 weeks old | "DCL1a mutant, Control" | none |
| 56 | SRR6498707 | SINGLE | leaf | 3 weeks old | "DCL1a mutant, M. oryzae Infected" | none |
| 57 | SRR6498708 | SINGLE | leaf | 3 weeks old | "WT, Control" | none |
| 58 | SRR6498709 | SINGLE | leaf | 3 weeks old | "WT, M. oryzae Infected" | none |
| 59 | SRR6498710 | SINGLE | leaf | 3 weeks old | "WT, Control" | none |
| 60 | SRR6498711 | SINGLE | leaf | 3 weeks old | "WT, M. oryzae Infected" | none |
| 61 | SRR1743007 | PAIRED | endosperm | 6 day after pollination | 4X_En_RNA | none |
| 62 | SRR1743008 | PAIRED | endosperm | 6 day after pollination | 4X_En_RNA | none |
| 63 | SRR1743009 | PAIRED | endosperm | 6 day after pollination | 4X_En_RNA | none |
| 64 | SRR1743010 | PAIRED | endosperm | 6 day after pollination | 2X4_En_RNA | none |
| 65 | SRR1743011 | PAIRED | endosperm | 6 day after pollination | 2X4_En_RNA | none |
| 66 | SRR1743012 | PAIRED | endosperm | 6 day after pollination | 2X4_En_RNA | none |
| 67 | SRR1743013 | PAIRED | endosperm | 6 day after pollination | 4X2_En_RNA | none |
| 68 | SRR1743014 | PAIRED | endosperm | 6 day after pollination | 4X2_En_RNA | none |
| 69 | SRR1743015 | PAIRED | endosperm | 6 day after pollination | 4X2_En_RNA | none |
| 70 | SRR4098173 | PAIRED | seedling | 2 weeks | No treatment | japanica |
| 71 | SRR4098176 | PAIRED | seedling | 2 weeks | Cold (4C) | japanica |
| 72 | SRR4098179 | PAIRED | seedling | 2 weeks | Drought (air-dried) | japanica |
| 73 | SRR4098187 | PAIRED | seedling | 2 weeks | Highlight (50000 Lux) | japanica |
| 74 | SRR4098190 | PAIRED | seedling | 2 weeks | High-salt (300 mM NaCl) | japanica |
| 75 | SRR4098193 | PAIRED | panicle | 2 weeks | No treatment | japanica |
| 76 | SRR4098196 | PAIRED | root | 2 weeks | No treatment | japanica |
| 77 | SRR4098199 | PAIRED | leaf | 2 weeks | No treatment | japanica |
| 78 | SRR6807844 | PAIRED | flag_leaf | Four weeks old seedlings | flag_leaf | PB-1 |
| 79 | SRR6807845 | PAIRED | flag_leaf | Four weeks old seedlings | flag_leaf | PB-1 |
| 80 | SRR6807850 | PAIRED | panicle | Four weeks old seedlings | panicle | PB-1 |
| 81 | SRR6807851 | PAIRED | panicle | Four weeks old seedlings | panicle | PB-1 |
| 82 | SRR3203681 | SINGLE | leaf | two-weeks | WT | none |
| 83 | SRR3203682 | SINGLE | leaf | two-weeks | 307T1 | none |
| 84 | SRR3203683 | SINGLE | leaf | two-weeks | 307T3 | none |
| 85 | SRR2862232 | SINGLE | leaf | three-leaf stage | mock leaf cells | cv. Wuyujing 3 |
| 86 | SRR2862233 | SINGLE | leaf | three-leaf stage | mock leaf cells | cv. Wuyujing 3 |
| 87 | SRR2862234 | SINGLE | leaf | three-leaf stage | mock leaf cells | cv. Wuyujing 3 |
| 88 | SRR2862235 | SINGLE | leaf | three-leaf stage | viruliferous Infected leaf cells | cv. Wuyujing 3 |
| 89 | SRR2862236 | SINGLE | leaf | three-leaf stage | viruliferous Infected leaf cells | cv. Wuyujing 3 |
| 90 | SRR2862237 | SINGLE | leaf | three-leaf stage | viruliferous Infected leaf cells | cv. Wuyujing 3 |
| 91 | SRR3130386 | SINGLE | leaf | fully expanded third leaf | control 0 hour | Co39 |
| 92 | SRR3130387 | SINGLE | leaf | fully expanded third leaf | control 0 hour | C101LAC |
| 93 | SRR3130388 | SINGLE | leaf | fully expanded third leaf | 8 hpi | Co39 |
| 94 | SRR3130389 | SINGLE | leaf | fully expanded third leaf | 8 hpi | C101LAC |
| 95 | SRR3130390 | SINGLE | leaf | fully expanded third leaf | control 8 hour | Co39 |
| 96 | SRR3130391 | SINGLE | leaf | fully expanded third leaf | control 8 hour | C101LAC |
| 97 | SRR3130392 | SINGLE | leaf | fully expanded third leaf | 24 hpi | Co39 |
| 98 | SRR3130393 | SINGLE | leaf | fully expanded third leaf | 24 hpi | C101LAC |
| 99 | SRR3130394 | SINGLE | leaf | fully expanded third leaf | 48 hpi | Co39 |
| 100 | SRR3130395 | SINGLE | leaf | fully expanded third leaf | 48 hpi | C101LAC |
| 101 | SRR1013790 | SINGLE | flag_leaf | Grain-filling stage | PA64s_FL | PA64s |
| 102 | SRR1013791 | SINGLE | flag_leaf | Grain-filling stage | LYP9_FL | LYP9 |
| 103 | SRR1013792 | SINGLE | flag_leaf | Grain-filling stage | 93-11_FL | 9311 |
| 104 | SRR1013793 | SINGLE | panicle | Grain-filling stage | PA64s_PA | PA64s |
| 105 | SRR1013794 | SINGLE | panicle | Grain-filling stage | LYP9_PA | LYP9 |
| 106 | SRR1013795 | SINGLE | panicle | Grain-filling stage | 93-11_PA | 9311 |
| 107 | SRR976168 | SINGLE | lamina_joint | none | wild type | Nipponbare |
| 108 | SRR976169 | SINGLE | lamina_joint | none | OsDCL3aIR-1 (mutant) | Nipponbare |
| 109 | SRR976170 | SINGLE | lamina_joint | none | OsDCL3aIR-3 (mutant) | Nipponbare |
| 110 | SRR447117 | PAIRED | root | 14-day-old | control for As(III) 20/80 uM | Nipponbare |
| 111 | SRR447120 | PAIRED | root | 14-day-old | As(III) 20 uM | Nipponbare |
| 112 | SRR447122 | PAIRED | shoot | 14-day-old | control for As(III) 20/80 uM | Nipponbare |
| 113 | SRR447123 | PAIRED | shoot | 14-day-old | As(III) 20 uM | Nipponbare |
| 114 | SRR447124 | PAIRED | shoot | 14-day-old | As(III) 80 uM | Nipponbare |
| 115 | SRR447125 | PAIRED | shoot | 14-day-old | As(III) 20 uM | Nipponbare |
| 116 | SRR447126 | PAIRED | shoot | 14-day-old | As(III) 80 uM | Nipponbare |