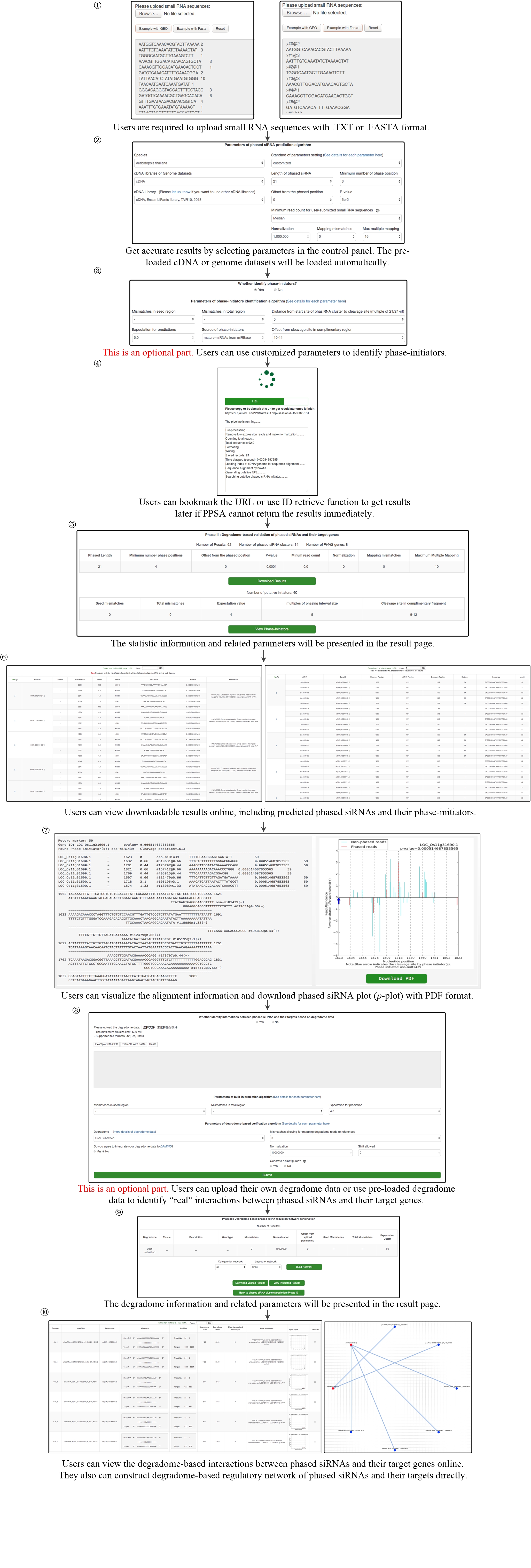
User manual
Fei Y, et al., 2021
Fei Y, et al., 2021

Fei Y, et al., 2021
| Parameters | Description |
|---|---|
| Standard of parameters setting | Set related parameters for phasiRNA predictions automatically, including customized, strict, normal and loose. |
| Phased length | Length of phasiRNA cluster, 21nt or 24 nt only |
| Minimum number of phased position | Minimum phased number in phasiRNA cluster |
| Offset from phased postion | Offset value from phased position, 0 or 1 only |
| P-value | The reliability of results, it calculated by improved hypergeometric distribution model |
| Minimum read count | Read count (expression) cutoff value for user-submitted small RNA sequences |
| Normalization | Normalization for user-submitted small RNA sequences, including 0 / Reads per million / Reads per 10 million |
| Mapping mismatches | Mismatches allowed for mapping small RNA sequences to either genomic or transcript datasets |
| Max multiple mapping | Remove multiple mapping that higher than this value |
Fei Y, et al., 2021
| Parameters | Description |
|---|---|
| Phase-initiator source | Candidate phase-initiators for identification. User-submitted small RNAs or mature-miRNAs come from miRBase |
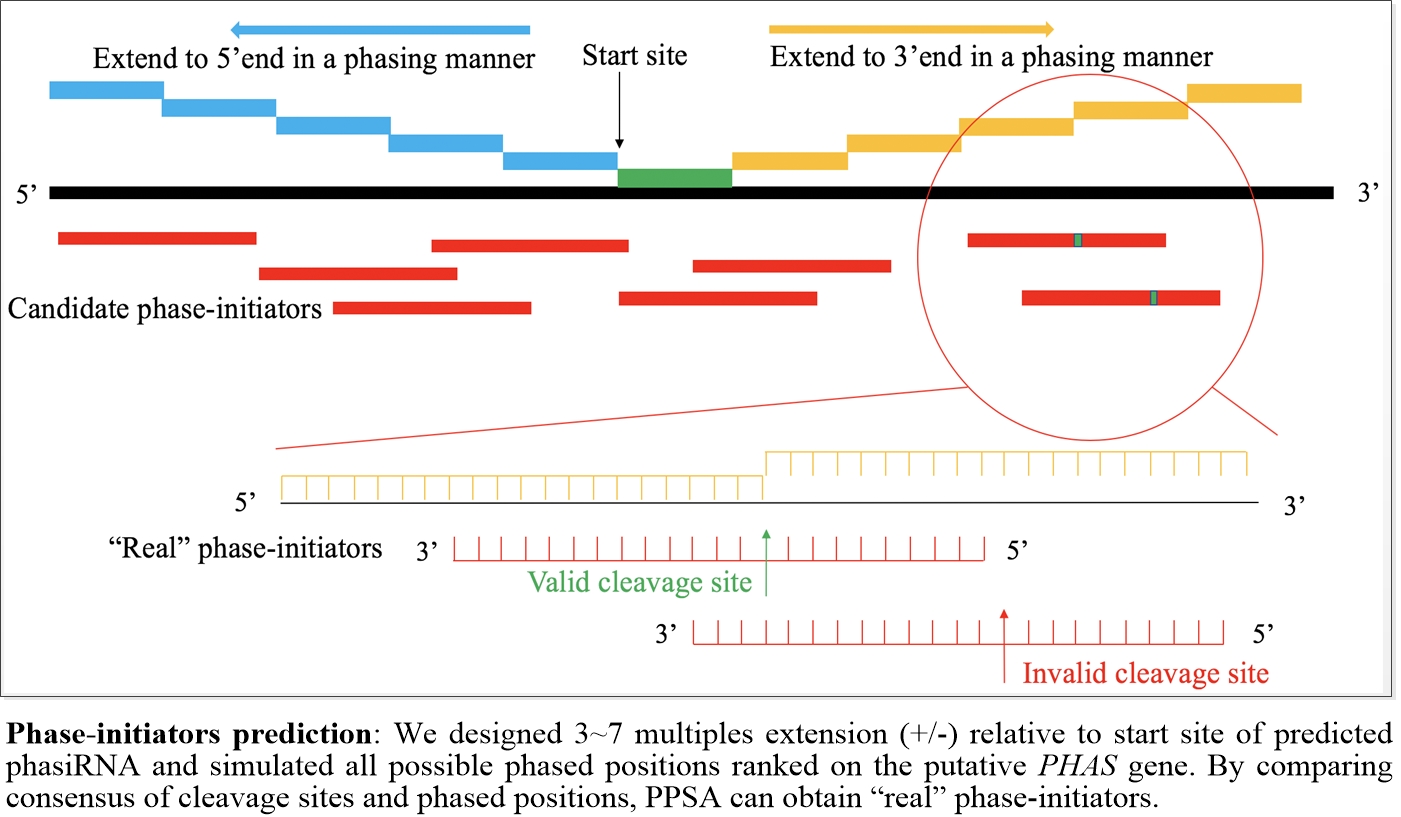
| Distance from start site of phasiRNA cluster to cleavage site | Max distance from cleavage site to phasiRNA clusters, (+/-) number * phased length (nt), number between 3 to 7 only |
| Offset from cleavage site in complimentary region | Cleavage site of complimentary region in candidate phase-initiators, 10th (0 offset) or 9~11th (1 offset) only |
| Other parameters | Seed mismatches, Total mismatches and Expectation value. |

Fei Y, et al., 2020
| Parameters | Description |
|---|---|
| Seed mismatches allowed | Mismatches happened in seed region (2~13bp) |
| Total mismatches allowed | Mismatches happened in total region |
| Expectation value for prediction | A lower value means more similarity between small RNA and the target mRNA |

Fei Y, et al., 2020
| Parameters | Description |
|---|---|
| Degradome | Degradome sequencing data for phasiRNA targets verification |
| Mismatches allowed for degradomes | Mismatches allowed for mapping degradome sequences to transcript (mRNA) |
| Normalization | Normalization for degradome sequencing data, including 0 / Reads per million / Reads per 10 million |
| Shift allowed | Cleavage site of complimentary region of phasiRNAs and target genes, 10th (0 offset) or 9~11th (1 offset) only |
| t-plot figures | Whether present t-plot figures in the result list |